Model Explainers - For Regression
Contents
Model Explainers - For Regression#
Lesson Objectives#
By the end of this lesson students will be able to:
Define a global vs local explanation
Use the Shap package and interpret shap values.
Model Explainers#
There are packages with the sole purpose of better understanding how machine learning models make their predictions.
Generally, model explainers will take the model and some of your data and apply some iterative process to try to quantify how the features are influencing the model’s output.
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
## Customization Options
# pd.set_option('display.float_format',lambda x: f"{x:,.4f}")
pd.set_option("display.max_columns",100)
plt.style.use(['fivethirtyeight','seaborn-talk'])
mpl.rcParams['figure.facecolor']='white'
## additional required imports
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.impute import SimpleImputer
from sklearn.compose import make_column_transformer, make_column_selector, ColumnTransformer
from sklearn.pipeline import make_pipeline, Pipeline
from sklearn import metrics
from sklearn.base import clone
## fixing random for lesson generation
SEED = 321
np.random.seed(SEED)
## Adding folder above to path
import os, sys
sys.path.append(os.path.abspath('../../'))
## Load stack_functions with autoreload turned on
%load_ext autoreload
%autoreload 2
from CODE import stack_functions as sf
from CODE import prisoner_project_functions as pf
def show_code(function):
import inspect
from IPython.display import display,Markdown, display_markdown
code = inspect.getsource(function)
md_txt = f"```python\n{code}\n```"
return display(Markdown(md_txt))
## Load in the student erformance data
url = "https://docs.google.com/spreadsheets/d/e/2PACX-1vS6xDKNpWkBBdhZSqepy48bXo55QnRv1Xy6tXTKYzZLMPjZozMfYhHQjAcC8uj9hQ/pub?output=xlsx"
df = pd.read_excel(url,sheet_name='student-mat')
# df.drop(columns=['G1','G2'])
df.info()
df.head()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 395 entries, 0 to 394
Data columns (total 33 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 school 395 non-null object
1 sex 395 non-null object
2 age 395 non-null float64
3 address 395 non-null object
4 famsize 395 non-null object
5 Pstatus 395 non-null object
6 Medu 395 non-null float64
7 Fedu 395 non-null float64
8 Mjob 395 non-null object
9 Fjob 395 non-null object
10 reason 395 non-null object
11 guardian 395 non-null object
12 traveltime 395 non-null float64
13 studytime 395 non-null float64
14 failures 395 non-null float64
15 schoolsup 395 non-null object
16 famsup 395 non-null object
17 paid 395 non-null object
18 activities 395 non-null object
19 nursery 395 non-null object
20 higher 395 non-null object
21 internet 395 non-null object
22 romantic 395 non-null object
23 famrel 395 non-null float64
24 freetime 395 non-null float64
25 goout 395 non-null float64
26 Dalc 395 non-null float64
27 Walc 395 non-null float64
28 health 395 non-null float64
29 absences 395 non-null float64
30 G1 395 non-null float64
31 G2 395 non-null float64
32 G3 395 non-null float64
dtypes: float64(16), object(17)
memory usage: 102.0+ KB
| school | sex | age | address | famsize | Pstatus | Medu | Fedu | Mjob | Fjob | reason | guardian | traveltime | studytime | failures | schoolsup | famsup | paid | activities | nursery | higher | internet | romantic | famrel | freetime | goout | Dalc | Walc | health | absences | G1 | G2 | G3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GP | F | 18.0 | U | GT3 | A | 4.0 | 4.0 | at_home | teacher | course | mother | 2.0 | 2.0 | 0.0 | yes | no | no | no | yes | yes | no | no | 4.0 | 3.0 | 4.0 | 1.0 | 1.0 | 3.0 | 6.0 | 5.0 | 6.0 | 6.0 |
| 1 | GP | F | 17.0 | U | GT3 | T | 1.0 | 1.0 | at_home | other | course | father | 1.0 | 2.0 | 0.0 | no | yes | no | no | no | yes | yes | no | 5.0 | 3.0 | 3.0 | 1.0 | 1.0 | 3.0 | 4.0 | 5.0 | 5.0 | 6.0 |
| 2 | GP | F | 15.0 | U | LE3 | T | 1.0 | 1.0 | at_home | other | other | mother | 1.0 | 2.0 | 3.0 | yes | no | yes | no | yes | yes | yes | no | 4.0 | 3.0 | 2.0 | 2.0 | 3.0 | 3.0 | 10.0 | 7.0 | 8.0 | 10.0 |
| 3 | GP | F | 15.0 | U | GT3 | T | 4.0 | 2.0 | health | services | home | mother | 1.0 | 3.0 | 0.0 | no | yes | yes | yes | yes | yes | yes | yes | 3.0 | 2.0 | 2.0 | 1.0 | 1.0 | 5.0 | 2.0 | 15.0 | 14.0 | 15.0 |
| 4 | GP | F | 16.0 | U | GT3 | T | 3.0 | 3.0 | other | other | home | father | 1.0 | 2.0 | 0.0 | no | yes | yes | no | yes | yes | no | no | 4.0 | 3.0 | 2.0 | 1.0 | 2.0 | 5.0 | 4.0 | 6.0 | 10.0 | 10.0 |
# ### Train Test Split
## Make x and y variables
drop_feats = ['G1','G2']
y = df['G3'].copy()
X = df.drop(columns=['G3',*drop_feats]).copy()
## train-test-split with random state for reproducibility
X_train, X_test, y_train, y_test = train_test_split(X,y, random_state=SEED)
# ### Preprocessing + ColumnTransformer
## make categorical & numeric selectors
cat_sel = make_column_selector(dtype_include='object')
num_sel = make_column_selector(dtype_include='number')
## make pipelines for categorical vs numeric data
cat_pipe = make_pipeline(SimpleImputer(strategy='constant',
fill_value='MISSING'),
OneHotEncoder(drop='if_binary', sparse=False))
num_pipe = make_pipeline(SimpleImputer(strategy='mean'))
## make the preprocessing column transformer
preprocessor = make_column_transformer((num_pipe, num_sel),
(cat_pipe,cat_sel),
verbose_feature_names_out=False)
## fit column transformer and run get_feature_names_out
preprocessor.fit(X_train)
feature_names = preprocessor.get_feature_names_out()
X_train_df = pd.DataFrame(preprocessor.transform(X_train),
columns = feature_names, index = X_train.index)
X_test_df = pd.DataFrame(preprocessor.transform(X_test),
columns = feature_names, index = X_test.index)
X_test_df.head(3)
| age | Medu | Fedu | traveltime | studytime | failures | famrel | freetime | goout | Dalc | Walc | health | absences | school_MS | sex_M | address_U | famsize_LE3 | Pstatus_T | Mjob_at_home | Mjob_health | Mjob_other | Mjob_services | Mjob_teacher | Fjob_at_home | Fjob_health | Fjob_other | Fjob_services | Fjob_teacher | reason_course | reason_home | reason_other | reason_reputation | guardian_father | guardian_mother | guardian_other | schoolsup_yes | famsup_yes | paid_yes | activities_yes | nursery_yes | higher_yes | internet_yes | romantic_yes | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 58 | 15.0 | 1.0 | 2.0 | 1.0 | 2.0 | 0.0 | 4.0 | 3.0 | 2.0 | 1.0 | 1.0 | 5.0 | 2.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 338 | 18.0 | 3.0 | 3.0 | 1.0 | 4.0 | 0.0 | 5.0 | 3.0 | 3.0 | 1.0 | 1.0 | 1.0 | 7.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 291 | 17.0 | 4.0 | 3.0 | 1.0 | 3.0 | 0.0 | 4.0 | 2.0 | 2.0 | 1.0 | 2.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 |
## fit random fores
from sklearn.ensemble import RandomForestRegressor
rf_reg = RandomForestRegressor()
rf_reg.fit(X_train_df,y_train)
sf.evaluate_regression(rf_reg,X_test_df,y_test, warn=False,
X_train=X_train_df, y_train=y_train)#linreg(rf_reg,X_train_zips,y_train,X_test_zips,y_test)
Train Test Delta
R^2 0.910 0.085 -0.825
RMSE 1.405 4.023 2.618
Fjob_health 0.001351
guardian_mother 0.004300
reason_other 0.004932
guardian_father 0.005284
reason_home 0.005356
internet_yes 0.005659
Pstatus_T 0.006196
paid_yes 0.007169
address_U 0.007630
Mjob_health 0.007761
Mjob_other 0.007970
famsize_LE3 0.008351
Fjob_services 0.008621
Mjob_teacher 0.009024
Dalc 0.009191
school_MS 0.009252
nursery_yes 0.009272
Fjob_other 0.009453
Mjob_services 0.010012
activities_yes 0.010288
Fjob_teacher 0.010371
guardian_other 0.010720
Fjob_at_home 0.010922
romantic_yes 0.012431
famsup_yes 0.013015
higher_yes 0.014331
reason_reputation 0.015734
Fedu 0.020560
sex_M 0.021505
famrel 0.021921
reason_course 0.022823
Mjob_at_home 0.023137
schoolsup_yes 0.023192
Medu 0.024457
age 0.027299
traveltime 0.027808
health 0.031356
freetime 0.035128
Walc 0.036055
studytime 0.038027
goout 0.047714
failures 0.151020
absences 0.213404
Name: Feature Importance, dtype: float64
Loading Joblib of Regressions from Lesson 04#
import joblib
## If showing joblib in prior lesson, this cell will be included and further explained
loaded_data = joblib.load("../3_Feature_Importance/lesson_03.joblib")
loaded_data.keys()
dict_keys(['X_train', 'y_train', 'X_test', 'y_test', 'preprocessor', 'lin_reg', 'rf_reg'])
## If showing joblib in prior lesson, this cell will be included and further explained
X_train_reg = loaded_data['X_train'].copy()
y_train_reg = loaded_data['y_train'].copy()
X_test_df_reg = loaded_data['X_test'].copy()
y_test_reg = loaded_data['y_test'].copy()
lin_reg = loaded_data['lin_reg']
rf_reg = loaded_data['rf_reg']
Using SHAP for Model Interpretation#
SHAP (SHapley Additive exPlanations))
SHAP uses game theory to calcualte Shapely values for each feature in the dataset.
Shapely values are calculated by iteratively testing each feature’s contribution to the model by comparing the model’s performance with vs. without the feature. (The “marginal contribution” of the feature to the model’s performance).
Papers, Book Excerpts, and Blogs#
Videos/Talks:#
Explaining Machine Learning Models (in general).
Understanding Shapely/SHAP Values:
AI Simplified: SHAP Values in Machine Learning - (Intuitive Explanation)
Explainable AI explained! | #4 SHAP - (Math Calculation Explanation)
How To Use Shap#
Import and initialize javascript:
import shap
shap.initjs()
import shap
shap.initjs()
Shap Explainers#
shap has several types of model explainers that are optimized for different types of models.
Explainers and their use cases:#
Explainer |
Description |
|---|---|
shap.Explainer |
Uses Shapley values to explain any machine learning model or python function. |
shap.explainers.Tree |
Uses Tree SHAP algorithms to explain the output of ensemble tree models. |
shap.explainers.Linear |
Computes SHAP values for a linear model, optionally accounting for inter-feature correlations. |
shap.explainers.Permutation |
This method approximates the Shapley values by iterating through permutations of the inputs. |
See this blog post for intro to topic and how to use with trees
For non-tree/random forest models see this follow up post
Preparing Data for Shap#
Shap’s approach to explaining models can be very resource-intensive for complex models such as our RandomForest.
To get around this issue, shap includes a convenient smapling function to save a small sample from one of our X variables.
X_shap = shap.sample(X_train_df,nsamples=200,random_state=321)
X_shap
| age | Medu | Fedu | traveltime | studytime | failures | famrel | freetime | goout | Dalc | Walc | health | absences | school_MS | sex_M | address_U | famsize_LE3 | Pstatus_T | Mjob_at_home | Mjob_health | Mjob_other | Mjob_services | Mjob_teacher | Fjob_at_home | Fjob_health | Fjob_other | Fjob_services | Fjob_teacher | reason_course | reason_home | reason_other | reason_reputation | guardian_father | guardian_mother | guardian_other | schoolsup_yes | famsup_yes | paid_yes | activities_yes | nursery_yes | higher_yes | internet_yes | romantic_yes | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 355 | 18.0 | 3.0 | 3.0 | 1.0 | 2.0 | 0.0 | 5.0 | 3.0 | 4.0 | 1.0 | 1.0 | 5.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 |
| 354 | 17.0 | 4.0 | 3.0 | 2.0 | 2.0 | 0.0 | 4.0 | 5.0 | 5.0 | 1.0 | 3.0 | 2.0 | 4.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 |
| 328 | 17.0 | 4.0 | 4.0 | 1.0 | 3.0 | 0.0 | 5.0 | 4.0 | 4.0 | 1.0 | 3.0 | 4.0 | 7.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 231 | 17.0 | 2.0 | 2.0 | 2.0 | 2.0 | 0.0 | 4.0 | 5.0 | 2.0 | 1.0 | 1.0 | 1.0 | 4.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 312 | 19.0 | 1.0 | 2.0 | 1.0 | 2.0 | 1.0 | 4.0 | 5.0 | 2.0 | 2.0 | 2.0 | 4.0 | 3.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 210 | 19.0 | 3.0 | 3.0 | 1.0 | 4.0 | 0.0 | 4.0 | 3.0 | 3.0 | 1.0 | 2.0 | 3.0 | 10.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 236 | 17.0 | 2.0 | 2.0 | 1.0 | 2.0 | 0.0 | 4.0 | 4.0 | 2.0 | 5.0 | 5.0 | 4.0 | 4.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 |
| 10 | 15.0 | 4.0 | 4.0 | 1.0 | 2.0 | 0.0 | 3.0 | 3.0 | 3.0 | 1.0 | 2.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 13 | 15.0 | 4.0 | 3.0 | 2.0 | 2.0 | 0.0 | 5.0 | 4.0 | 3.0 | 1.0 | 2.0 | 3.0 | 2.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 372 | 17.0 | 2.0 | 2.0 | 1.0 | 3.0 | 0.0 | 3.0 | 4.0 | 3.0 | 1.0 | 1.0 | 3.0 | 8.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 |
200 rows × 43 columns
## get the corresponding y-values
y_shap = y_train.loc[X_shap.index]
y_shap
355 9.0
354 11.0
328 9.0
231 11.0
312 11.0
...
210 8.0
236 13.0
10 9.0
13 11.0
372 11.0
Name: G3, Length: 200, dtype: float64
Explaining Our RandomForest#
Create a shap explainer using your fit model.
explainer = shap.TreeExplainer(rf_reg)
Get shapely values from explainer for your training data
shap_values = explainer(X_shap)
Select which type of the available plots you’d like to visualize
Types of Plots:
summary_plot()dependence_plot()force_plot()for a given observationforce_plot()for all data
X_shap = X_train_df.copy()#shap.sample(X_train_df,nsamples=200,random_state=SEED)
explainer = shap.TreeExplainer(rf_reg)
shap_values = explainer(X_shap,y_shap)
shap_values[0]
.values =
array([-2.65129202e-03, 3.37739374e-02, -1.66913694e-02, 4.04224947e-02,
-8.89278699e-02, 8.22963912e-01, -2.19713066e-02, 9.46616336e-03,
-1.05073602e-01, 4.45105221e-02, 7.95999848e-02, 6.99744889e-01,
1.05407076e+00, 7.37235898e-03, -1.65178625e-01, 1.09019192e-01,
6.88803146e-02, -8.73117299e-03, 1.44210492e-01, -4.05021571e-02,
8.44343694e-03, -5.94057998e-02, 9.03834608e-02, 2.51376396e-02,
1.69540791e-03, -5.10195738e-02, -2.51987310e-03, -3.91344713e-02,
1.97009755e-01, -3.56228354e-03, -2.04581060e-02, 2.76732166e-01,
-1.81765629e-03, 1.43730639e-02, 5.16251179e-02, 1.98845113e-01,
2.61826118e-01, -5.57521403e-04, -2.41598627e-02, 3.22229694e-02,
9.33188335e-03, 3.45140981e-02, 7.30116122e-02])
.base_values =
array([10.34317568])
.data =
array([17., 3., 2., 2., 2., 0., 4., 4., 4., 1., 3., 1., 2.,
0., 0., 1., 1., 1., 0., 0., 1., 0., 0., 0., 0., 1.,
0., 0., 0., 0., 0., 1., 0., 1., 0., 0., 0., 1., 0.,
1., 1., 1., 0.])
X_shap.shape
(296, 43)
shap_values.shape
(296, 43)
We can see that shap calculated values for every row/column in our X_shap variable.
What does the first row’s shap values look like?
shap_values[0]
.values =
array([-2.65129202e-03, 3.37739374e-02, -1.66913694e-02, 4.04224947e-02,
-8.89278699e-02, 8.22963912e-01, -2.19713066e-02, 9.46616336e-03,
-1.05073602e-01, 4.45105221e-02, 7.95999848e-02, 6.99744889e-01,
1.05407076e+00, 7.37235898e-03, -1.65178625e-01, 1.09019192e-01,
6.88803146e-02, -8.73117299e-03, 1.44210492e-01, -4.05021571e-02,
8.44343694e-03, -5.94057998e-02, 9.03834608e-02, 2.51376396e-02,
1.69540791e-03, -5.10195738e-02, -2.51987310e-03, -3.91344713e-02,
1.97009755e-01, -3.56228354e-03, -2.04581060e-02, 2.76732166e-01,
-1.81765629e-03, 1.43730639e-02, 5.16251179e-02, 1.98845113e-01,
2.61826118e-01, -5.57521403e-04, -2.41598627e-02, 3.22229694e-02,
9.33188335e-03, 3.45140981e-02, 7.30116122e-02])
.base_values =
array([10.34317568])
.data =
array([17., 3., 2., 2., 2., 0., 4., 4., 4., 1., 3., 1., 2.,
0., 0., 1., 1., 1., 0., 0., 1., 0., 0., 0., 0., 1.,
0., 0., 0., 0., 0., 1., 0., 1., 0., 0., 0., 1., 0.,
1., 1., 1., 0.])
Notice above that we do not seem to have a simple numpy array.
type(shap_values[0])
shap._explanation.Explanation
explanation_0 = shap_values[0]
explanation_0
.values =
array([-2.65129202e-03, 3.37739374e-02, -1.66913694e-02, 4.04224947e-02,
-8.89278699e-02, 8.22963912e-01, -2.19713066e-02, 9.46616336e-03,
-1.05073602e-01, 4.45105221e-02, 7.95999848e-02, 6.99744889e-01,
1.05407076e+00, 7.37235898e-03, -1.65178625e-01, 1.09019192e-01,
6.88803146e-02, -8.73117299e-03, 1.44210492e-01, -4.05021571e-02,
8.44343694e-03, -5.94057998e-02, 9.03834608e-02, 2.51376396e-02,
1.69540791e-03, -5.10195738e-02, -2.51987310e-03, -3.91344713e-02,
1.97009755e-01, -3.56228354e-03, -2.04581060e-02, 2.76732166e-01,
-1.81765629e-03, 1.43730639e-02, 5.16251179e-02, 1.98845113e-01,
2.61826118e-01, -5.57521403e-04, -2.41598627e-02, 3.22229694e-02,
9.33188335e-03, 3.45140981e-02, 7.30116122e-02])
.base_values =
array([10.34317568])
.data =
array([17., 3., 2., 2., 2., 0., 4., 4., 4., 1., 3., 1., 2.,
0., 0., 1., 1., 1., 0., 0., 1., 0., 0., 0., 0., 1.,
0., 0., 0., 0., 0., 1., 0., 1., 0., 0., 0., 1., 0.,
1., 1., 1., 0.])
Each entry in the shap_values array is new type of object called an Explanation.
Each Explanation has:
values:the shap values calculated for this observation/row.
For classification models, there is a column with values for each target.
base_values: the final shap output value
data: the original input feature
## Showing .data is the same as the raw X_shap
explanation_0.data
array([17., 3., 2., 2., 2., 0., 4., 4., 4., 1., 3., 1., 2.,
0., 0., 1., 1., 1., 0., 0., 1., 0., 0., 0., 0., 1.,
0., 0., 0., 0., 0., 1., 0., 1., 0., 0., 0., 1., 0.,
1., 1., 1., 0.])
X_shap.iloc[0].values
array([17., 3., 2., 2., 2., 0., 4., 4., 4., 1., 3., 1., 2.,
0., 0., 1., 1., 1., 0., 0., 1., 0., 0., 0., 0., 1.,
0., 0., 0., 0., 0., 1., 0., 1., 0., 0., 0., 1., 0.,
1., 1., 1., 0.])
## showing the .values
pd.Series(explanation_0.values,index=X_shap.columns)
age -0.002651
Medu 0.033774
Fedu -0.016691
traveltime 0.040422
studytime -0.088928
failures 0.822964
famrel -0.021971
freetime 0.009466
goout -0.105074
Dalc 0.044511
Walc 0.079600
health 0.699745
absences 1.054071
school_MS 0.007372
sex_M -0.165179
address_U 0.109019
famsize_LE3 0.068880
Pstatus_T -0.008731
Mjob_at_home 0.144210
Mjob_health -0.040502
Mjob_other 0.008443
Mjob_services -0.059406
Mjob_teacher 0.090383
Fjob_at_home 0.025138
Fjob_health 0.001695
Fjob_other -0.051020
Fjob_services -0.002520
Fjob_teacher -0.039134
reason_course 0.197010
reason_home -0.003562
reason_other -0.020458
reason_reputation 0.276732
guardian_father -0.001818
guardian_mother 0.014373
guardian_other 0.051625
schoolsup_yes 0.198845
famsup_yes 0.261826
paid_yes -0.000558
activities_yes -0.024160
nursery_yes 0.032223
higher_yes 0.009332
internet_yes 0.034514
romantic_yes 0.073012
dtype: float64
Shap Visualizations - Regression#
Summary Plot#
## For normal bar graph of importance:
shap.summary_plot(shap_values, features=X_shap, plot_type='bar')
## For detail Shapely value visuals:
shap.summary_plot(shap_values, features=X_shap)
shap.summary_plot
Feature importance: Variables are ranked in descending order.
Impact: The horizontal location shows whether the effect of that value is associated with a higher or lower prediction.
Original value: Color shows whether that variable is high (in red) or low (in blue) for that observation.
IMPORTANT NOTE: You may need to slice out the correct shap_values for the target class. (by default explainer.shap_values seems to return a list for a binary classification, one set of shap values for each class). - This will cause issues like the summary plot having a bar with an equal amount of blue and red for each class. - To fix, slice out the correct matrix from shap_values [0,1]
First, let’s examine a simple version of our shap values.
By using the plot_type=”bar” version of the summary plot, we get something that looks very similar to the feature importances we discussed previously.
shap.summary_plot(shap_values,features= X_shap,plot_type='bar')
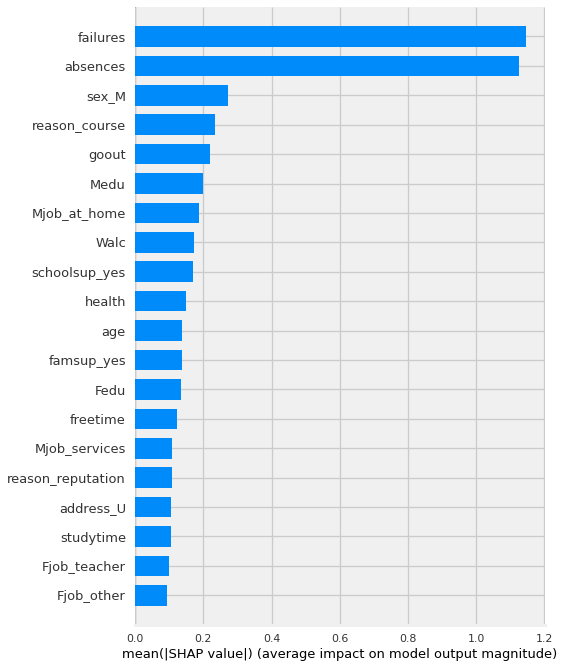
In this case, it is using the magnitude of the average shap values to to show which features had the biggest impact on the model’s predictions.
Like feature importance and permutation importance, this visualization is not indicating which direction the features push the predict.
Now, let’s examine the “dot” version of the summary plot.
By removing the plot_type argument, we are using the default, which is “dot”.
We could explicitly specify plot_type=”dot”.
There are also additional plot types that we will not be discussing in this lesson (e.g. “violin”,”compact_dot”)
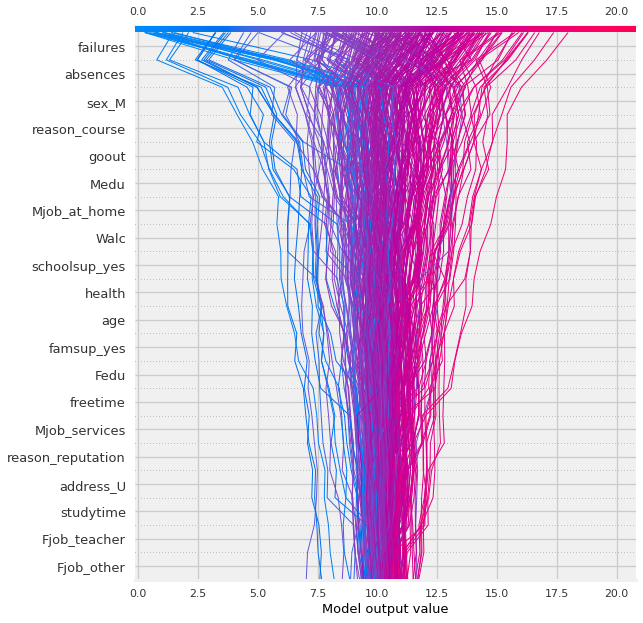
shap.summary_plot(shap_values,X_shap)
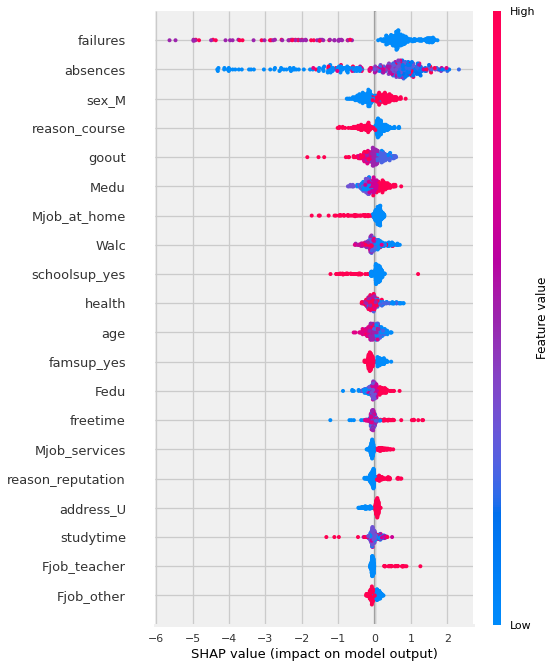
Now THAT is a lot more nuanced of a visualization! Let’s break down how to interpret the visual above.
Reading Summary Plots#
In the summary plot above:
Each dot represents an observation/row (in this case, a student).
The features are plotting on the y-axis, and are sorted from the most impactful features to the least (from top to bottom).
The calculated Shapely values for each observation are plotted on the x-axis. The most positive the value the more positive…
bookmarkFor each feature, the original values of that feature are represented with color.
Using the default colormap, blue represents the lowest value in the column and red represents the highest.
For one hot encoded categories, blue=0, red = 1.
For numeric features: the shade of the color indicates where it falls in the feature’s distribution.
Summary Plot Interpretation#
fewer prior failures = higher final grade -
Q: what is going on with absences?why are some of the lowest values leading to negative shap value (meaning a decreased final score)?
Why would less absences meaning a lower final grade?
🤔 Did the student not spend all 3 years at the school??
sex_M:
males get a higher grade
reason_course:
if a student attends the school because of a specific course, they have a lower final grade.
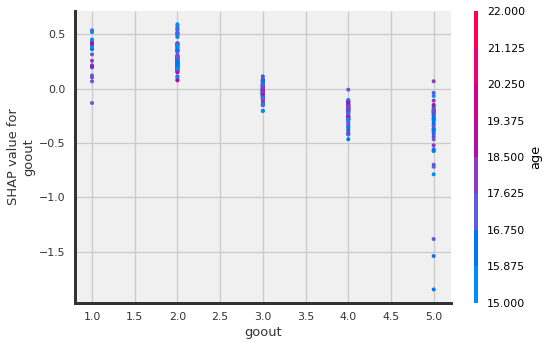
goout:
the more a student goes out, the lower the grade.
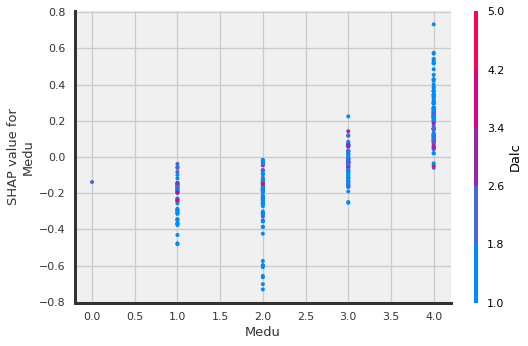
Medu (mother’s education):
Higher mother’s ed the higher the grade
Mjob_at_home:
Mother at home leads to lower grades.
Walc: Lower weekend alcohol consumption “causes” higher grade
Dependence Plots#
Shap also includes the shap.dependence_plot
which show how the model output varies by a specific feature. By passing the function a feature name, it will automatically determine what features may driving the interactions with the selected feature. It will encode the interaction feature as color.
## To Auto-Select Feature Most correlated with a specific feature, just pass the desired feature's column name.
shap.dependence_plot('age', shap_values, X_shap)
TO DO:
There is a way to specifically call out multiple features but I wasn’t able to summarize it quickly for this nb
## Using shap_values made from shap_values = explainer(X_shap)
shap.dependence_plot("absences", shap_values.values,X_shap)
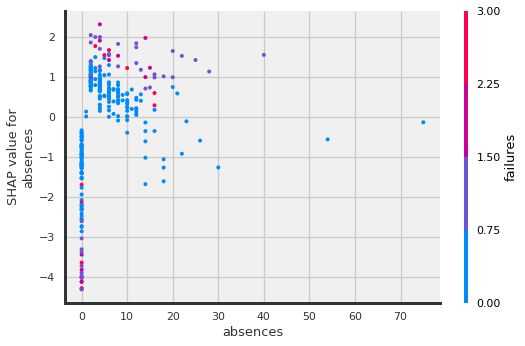
## Using shap_values made from shap_values = explainer(X_shap)
shap.dependence_plot("Medu", shap_values.values,X_shap)
## Using shap_values made from shap_values = explainer(X_shap)
shap.dependence_plot("goout", shap_values.values,X_shap)
Force Plot#
Note: the force_plot is an interactive visualization that uses javascript. You must Trust your jupyter notebook in order to display it. - In the top right corner of jupyter notebook, next the kernel name (Python (dojo-env)), click the
Not Trustedbutton to trust the notebook.
Global shap.force_plot#
To show a global force plot:
## Fore plot
shap.force_plot(explainer.expected_value[1], shap_values[:,:,1], features=X_shap)
Global Force Plot#
## TESTING COMPLEX SHAP VALS AGAIN (Overall Forceplot)
shap.force_plot(explainer.expected_value, shap_values.values,features=X_shap)
Have you run `initjs()` in this notebook? If this notebook was from another user you must also trust this notebook (File -> Trust notebook). If you are viewing this notebook on github the Javascript has been stripped for security. If you are using JupyterLab this error is because a JupyterLab extension has not yet been written.
Fore Plot Interpretation#
TO DO
Explain Individual Plot#
To show an individual data point’s prediction and the factors pushing it towards one class or another.
For now, we will randomly select a row to display, but we will revisit thoughtful selection of examples for stakeholders in our next lesson about local explanations.
## Just using np to randomly select a row
row = np.random.choice(range(len(X_shap)))
shap.force_plot(explainer.expected_value[1], shap_values[1][row], X_shap.iloc[row])
row = np.random.choice(range(len(X_shap)))
print(f"- Row #: {row}")
print(f"- Target: {y_shap.iloc[row]}")
X_shap.iloc[row].round(2)
- Row #: 85
- Target: 6.0
age 18.0
Medu 1.0
Fedu 1.0
traveltime 2.0
studytime 2.0
failures 1.0
famrel 4.0
freetime 4.0
goout 3.0
Dalc 2.0
Walc 3.0
health 5.0
absences 2.0
school_MS 1.0
sex_M 1.0
address_U 0.0
famsize_LE3 1.0
Pstatus_T 1.0
Mjob_at_home 1.0
Mjob_health 0.0
Mjob_other 0.0
Mjob_services 0.0
Mjob_teacher 0.0
Fjob_at_home 0.0
Fjob_health 0.0
Fjob_other 1.0
Fjob_services 0.0
Fjob_teacher 0.0
reason_course 0.0
reason_home 0.0
reason_other 1.0
reason_reputation 0.0
guardian_father 0.0
guardian_mother 1.0
guardian_other 0.0
schoolsup_yes 0.0
famsup_yes 0.0
paid_yes 0.0
activities_yes 1.0
nursery_yes 0.0
higher_yes 0.0
internet_yes 0.0
romantic_yes 0.0
Name: 361, dtype: float64
## Individual forceplot (with the complex shap vals)
shap.force_plot(explainer.expected_value,shap_values= shap_values[row].values,
features=X_shap.iloc[row])
Have you run `initjs()` in this notebook? If this notebook was from another user you must also trust this notebook (File -> Trust notebook). If you are viewing this notebook on github the Javascript has been stripped for security. If you are using JupyterLab this error is because a JupyterLab extension has not yet been written.
Waterfall Plot#
explainer.expected_value
array([10.34317568])
shap_values[row]#,:,1]
.values =
array([-1.06826576e-01, -8.27475108e-02, -9.09468180e-02, -1.41339274e-02,
-5.23458955e-02, -1.40791878e+00, 6.95681721e-03, -4.14373232e-02,
-7.43381448e-02, -3.20156255e-02, -1.14419829e-01, -1.11251761e-01,
2.04968238e+00, 2.65195333e-02, 4.74230874e-02, -8.60920840e-02,
1.05248311e-01, -7.51729126e-03, 7.89352552e-02, -1.64113732e-02,
1.35993462e-01, -2.18647657e-02, 1.99391968e-02, -1.90337616e-02,
-1.55896265e-03, -3.25649041e-02, 2.07980786e-03, -2.77867960e-02,
9.32572748e-02, 1.00607556e-02, 1.00138176e-01, -1.92533952e-02,
3.49964031e-03, -4.32813322e-03, -1.97932944e-04, 3.07952569e-02,
9.92020595e-02, -8.22436248e-03, 6.61156139e-02, -1.49940694e-02,
-2.34602883e-01, 4.27374105e-02, 4.10531897e-02])
.base_values =
array([10.34317568])
.data =
array([18., 1., 1., 2., 2., 1., 4., 4., 3., 2., 3., 5., 2.,
1., 1., 0., 1., 1., 1., 0., 0., 0., 0., 0., 0., 1.,
0., 0., 0., 0., 1., 0., 0., 1., 0., 0., 0., 0., 1.,
0., 0., 0., 0.])
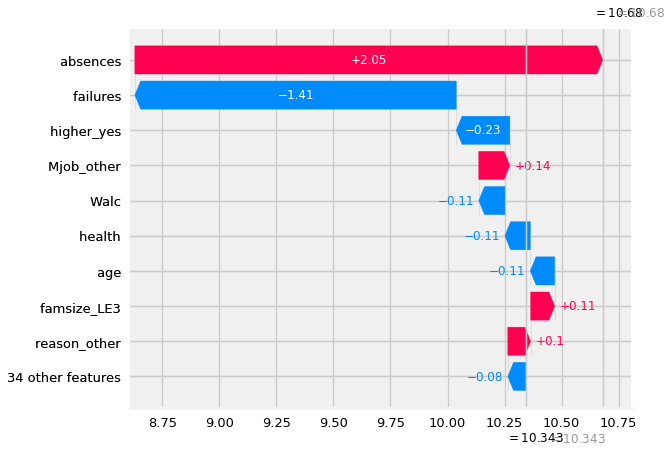
#source: https://towardsdatascience.com/explainable-ai-xai-a-guide-to-7-packages-in-python-to-explain-your-models-932967f0634b
shap.plots._waterfall.waterfall_legacy(explainer.expected_value[0],
shap_values[row].values,
features=X_shap.iloc[row],
show=True)
BOOKMARK: stopped here 07/11/22#
Interaction Values#
“The main effects are similar to the SHAP values you would get for a linear model, and the interaction effects captures all the higher-order interactions are divide them up among the pairwise interaction terms. Note that the sum of the entire interaction matrix is the difference between the model’s current output and expected output, and so the interaction effects on the off-diagonal are split in half (since there are two of each). When plotting interaction effects the SHAP package automatically multiplies the off-diagonal values by two to get the full interaction effect.”
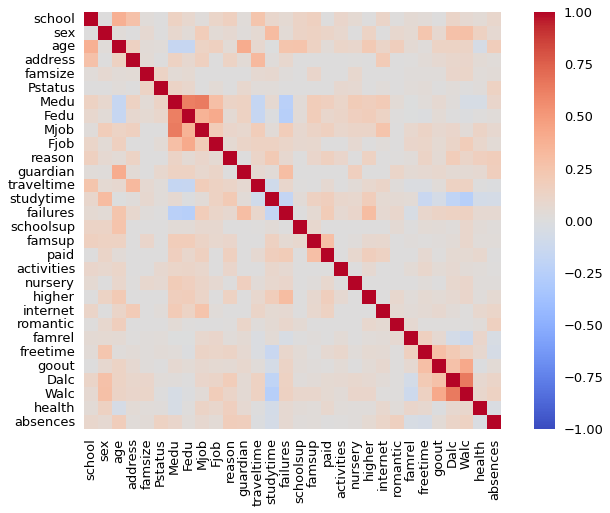
from dython.nominal import associations
res = associations(X, annot=False,cmap='coolwarm')
len(res)
2
shap_interaction_values = explainer.shap_interaction_values(X_shap)
shap.summary_plot(shap_interaction_values,X_shap)
shap.dependence_plot(
("age", "Walc"),
shap_interaction_values, X_shap,
display_features=X_shap
)
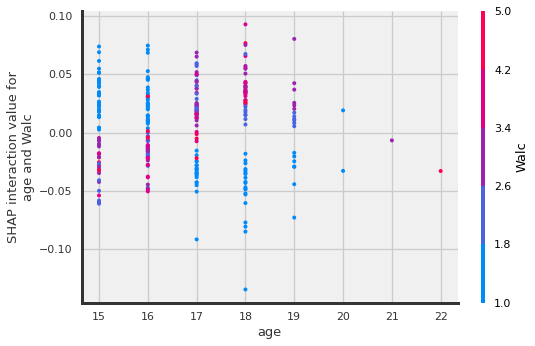
TO DO: read more about the interactions and add interpretation here
Shap Decision Plot?#
shap.decision_plot(explainer.expected_value, shap_values.values,X_shap)